Use of web services for building a gene tree
Gene trees (Goodman et al 1979) are staples of research in comparative biology (Hibbins et al 2023). Basics for building a gene tree with packages managed in UGENE were presented in Make a gene tree. Here, we describe use of NGPhylogeny web service. From the Lemoine et al (2019) abstract:
NGPhylogeny.fr … integrates numerous tools … [which] cover a large range of usage (sequence searching, multiple sequence alignment, model selection, tree inference and tree drawing) and a large panel of standard methods (distance, parsimony, maximum likelihood and Bayesian). They are integrated in workflows, which have been already configured (‘One click’), can be customized (‘Advanced’), or are built from scratch (‘A la carte’). Workflows are managed and run by an underlying Galaxy workflow system.
NGPhylogeny web service
Use of NGPhylogeny is straightforward. For example, select One click workflow, and select PhyML + SMS/One click from the menu. Assuming you have ready a file containing a list of protein or DNA sequences for a set of three or more taxa in FASTA format (e.g., use Batch Entrez), the workflow then is:
Input data → Alignment → Alignment curation → Tree inference → Tree rendering.
The only user input is to provide the sequence file. Alignment is by MAFFT, curation by BMGE, Tree inference by PhyML and SMS, ending with Tree rendering (building). All results files may be viewed or downloaded. Sequences may be viewed following alignment and alignment curation (MSAViewer) and run (Fig 1) ends with Newick files, also available for downloading or viewing in PRESTO (a tree viewer built on phylotree.js).
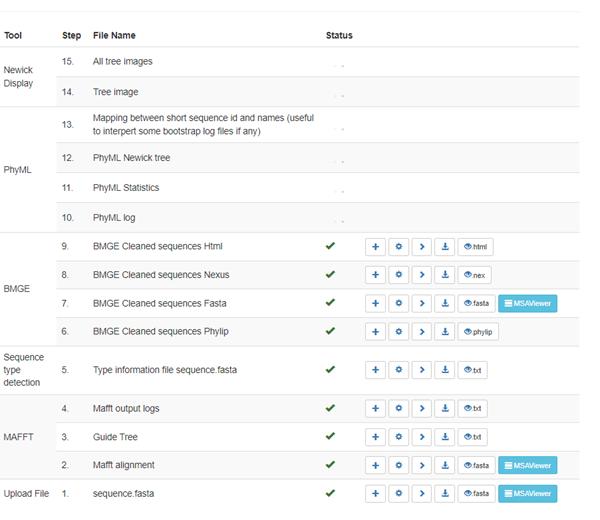
Figure 1. Screenshot of progress of NGPhylogeny run.
Complete documentation is available at https://ngphylogeny.fr/documentation .
References and suggested readings
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., … & Gascuel, O. (2008). Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research, 36(suppl_2), W465-W469. https://doi.org/10.1093/nar/gkn180
Goodman M, Czelusniak J, Moore G, Romero-Herrera A, Matsuda G: Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Systematic Zoology. 1979, 28: 132-163. https://doi.org/10.1093/sysbio/28.2.132
Hibbins, M. S., Breithaupt, L. C., & Hahn, M. W. (2023). Phylogenomic comparative methods: accurate evolutionary inferences in the presence of gene tree discordance. Proceedings of the National Academy of Sciences, 120(22), e2220389120. https://doi.org/10.1073/pnas.2220389120
Lefort, V., Longueville, J. E., & Gascuel, O. (2017). SMS: smart model selection in PhyML. Molecular biology and evolution, 34(9), 2422-2424. https://doi.org/10.1093/molbev/msx149
Lemoine, F., Correia, D., Lefort, V., Doppelt-Azeroual, O., Mareuil, F., Cohen-Boulakia, S., & Gascuel, O. (2019). NGPhylogeny. fr: new generation phylogenetic services for non-specialists. Nucleic acids research, 47(W1), W260-W265. https://doi.org/10.1093/nar/gkz303
Shank, S. D., Weaver, S., & Kosakovsky Pond, S. L. (2018). phylotree. js-a JavaScript library for application development and interactive data visualization in phylogenetics. BMC bioinformatics, 19, 1-5. https://doi.org/10.1186/s12859-018-2283-2