Download sequences with BATCH Entrez
Download and save multiple sequences in FASTA format from NCBI database.
You can download the sequences directly from NCBI protein. The advantage of this method is that you have the complete sequences which can then be imported to UGENE or any other bioinformatics app.
Batch implies a series of commands submitted to a local or remote computer to execute those commands without user intervention. The instructions on this page allow you to submit batch command to a NCBI database via Batch Entrez to recover multiple sequences with minimal user intervention
Workflow:
List of accessions → Go to BATCH Entrez → Choose database → upload file → Retrieve sequences → Save file
SOP
Build list of accessions
Acquire list of accessions for target sequences, Bioinformatics I: BLAST. For this example, common names of the taxa are
Alligator Cat Cattle Chicken Chimpanzee Dog Frog Human Lizard Macaque Mouse Opossum Pig Rabbit Rat
The input file is your list of accessions in a text file. For this example, my accessions are for 15 orthologous sequences of the protein HIF1A.
XP_059579361.1 XP_003987765 NP_776764 NP_989628 XP_009426180 NP_001274092 NP_001080449 NP_001521 XP_008121253 XP_005561497 NP_001300848 XP_007473044 NP_001116596 NP_001076251 NP_077335
Named the file hif1a_accessions.txt. Note the file ONLY has the accession numbers — nothing else! And, it must be a text only file (no formatting, no line spacing, etc.).
Go to Batch Entrez
https://www.ncbi.nlm.nih.gov/sites/batchentrez
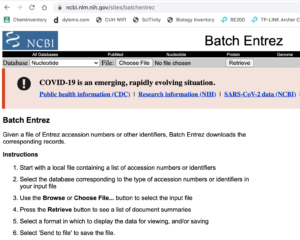
Figure 1. Screenshot of Batch Entrez homepage.
For this example, select Protein database.
Next, Choose file: locate the text file hif1a_accessions.txt(example), from your drive. Then, click on Retrieve button. If all goes well, a new page pops up with a message like Fig 2.

Figure 2. Screenshot of Batch Entrez results. It confirms that 15 of 15 accessions were found.
Click on link “Retrieve records” link. In this example, “Retrieve records of 15 UID(s)”. The results page lists the 15 sequences (Fig 3).
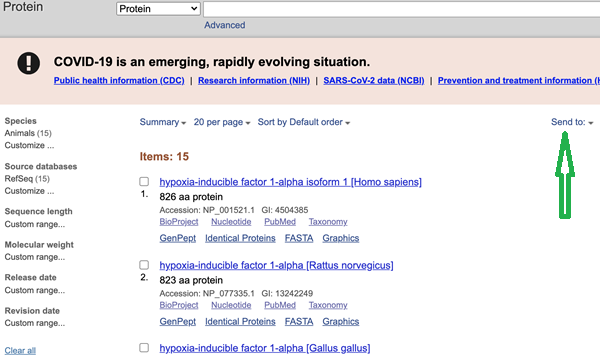
Figure 3. Screenshot web page of results listing 15 sequences by title.
Click on the down arrow next to “Send to” (highlighted by the green arrow I drew). The links are located to right and upper mid page in the browser window.
Choose File for destination and choose the FASTA format. Click “Create file” button when ready (Fig 4).
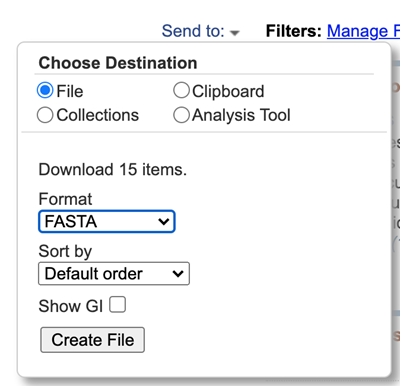
Figure 4. Screenshot of full Send to: menu options.
The file sequence.fasta will be located in your project Work folder.
Next, you should open the file in your text editor and change the NCBI sequence names to our 13+1 project common names. For example, NP_989628.2 from the list above corresponds to the protein sequence for Chicken. In the text file find:
>NP_989628.2 hypoxia-inducible factor 1-alpha isoform 1 [Gallus gallus]
and replace so the entry reads:
>Chicken
repeat to replace the other entries with proper common names for the project.