Use of ORF Finder: Practice and Worked example
This assignment supports Bioinformatics II
Purpose
Learn how to use ORF finder to identify candidate genes from DNA sequence.
Definition: ORF, the nominal gene, defined based on similarity to other known genes: Start/Stop, sufficient length of nucleotides (nt) to form a likely protein sequence. A putative gene, then is an ORF subsequently analyzed, eg, by bioinformatics methods like sequence similarities to known genes, and is strongly believed to be a functional gene.
What to do
Read through and try on your own to duplicate the results from the example sequence.
Try on your own — see bottom of this page, “What to do, what to turn in”
Resources
- link to ORF Finder, https://www.ncbi.nlm.nih.gov/orffinder/
- Use of UGENE.
- UGENE is a free, open-source bioinformatics platform for managing and analyzing biological sequences. Available at http://ugene.net/. See also Wikipedia entry.
Finding ORFs with ORF Finder
1. Do we have a gene? For this example, I generated a random sequence. Think of the random sequence as a null hypothesis: By searching multiple random sequences we can create a baseline for the expected number and length of ORFs that occur by chance alone. This null model helps determine the statistical significance of ORFs found in real biological sequences.
Copy/paste sequence in FASTA format to ORF Finder
Look for START (ATG) and STOP (TAG, TAA, or TGA) >random sequence 1 acagacctctattaccgttcccgatccacagggtggcggcattataaaccaacacgtacagccccccctctttactgcccacgaagcgcaccgtccctcgaatcgcggcgtctgcggaattgctatatttacagggcgtcaacaagcacacgcttcagacacgcttaaggtgtttccggcgtaccgaaaacgactgtttcctaggcgtgccgttcctagtaccttcgcttactgcaggtcccgatggcttgggagcggtagagatattctatcttgcttgcggttcgtcaacctatacataccgacatccgctgtcataccatcctcaaggaaagcctacattagttctatgcagccgcgtgagtaaaggggggagacgagagccctccggaagggatagcattacagagctgggggtacattaaacagctctcacagccactcacccatcgtcgaacggtggtcgtcctttccgtgaatcccgcaggctgttacggtctagagattcggtggagtagcactagggggtgtattgttaggtgagaggtacaaacgcttcaagtccctaatggattctcgctcgagagccggtcttcaccccgggctacaccgacggaactctcagcttctccaccatgatagcacccttgagcattcatctaatcaccgttttggtgggggataaggcttaaccgtccaagtcgccacagtccagtgcagtcggttaggatttggcgcaaattgggaagaaggcattccgatggttatacgagatatcagcaagtagatcgaagcggaatacgggtggtgtgtggaatgactacgcggaaagaacacgtcgcccatctgtagaaaaacacttttatatccctattaactgtgacattggattgtcgaggtcgtcgctcaagtcgtcaggtcattaatctaataacctggcgaataaccaggcgagcggataggcgatgcacaggacgtccttaccatgatatgatgccgt
ORF Finder screenshot
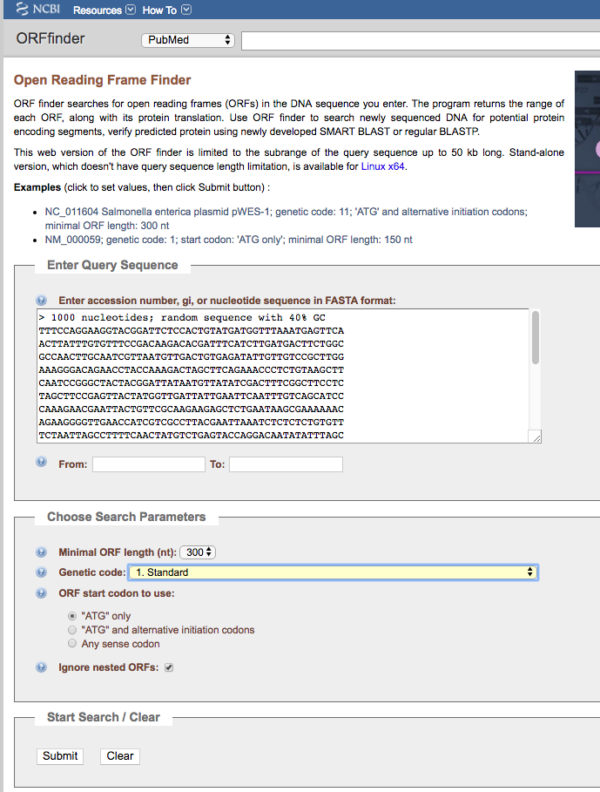
Figure 1. Screenshot of homepage of ORF Finder.
Note minimum ORF length was set to 300, the use of the standard genetic code, and only the ATG start codon was selected. We also elected to ignore nested ORFs (default). Unlike the main gene, internal ORFs can encode different proteins.
2. There are not a lot of options, but your direction should be based on a specific test.
Think strategically based on your definition of an ORF then approach the problem by adjusting settings in ORF finder to test.
For example, vary the number of nt and count returned ORFs.
The example on this page, shown in Fig 2, set ORF length to 30 nucleotides (nt). You could explore other genetic codes beside the standard — under what circumstances does it make sense to do this?
You should also consider whether or not your sequence should be evaluated as a nested gene, a gene sequence contained entirely within another gene. For example, microRNA genes are sometimes located in the introns of other protein coding genes.
Figure 2 provides screenshot of results from ORF finder.
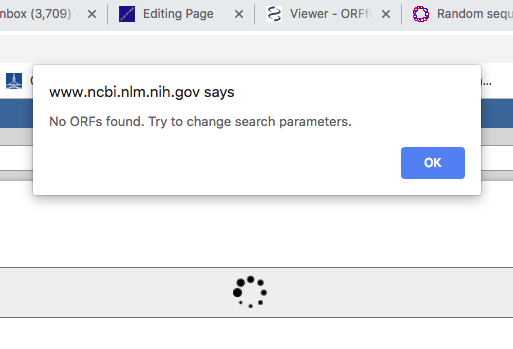
Figure 2 Screenshot of results from ORF finder on our random sequence 1.
Or, the second possibility, ORF finder does find an ORF
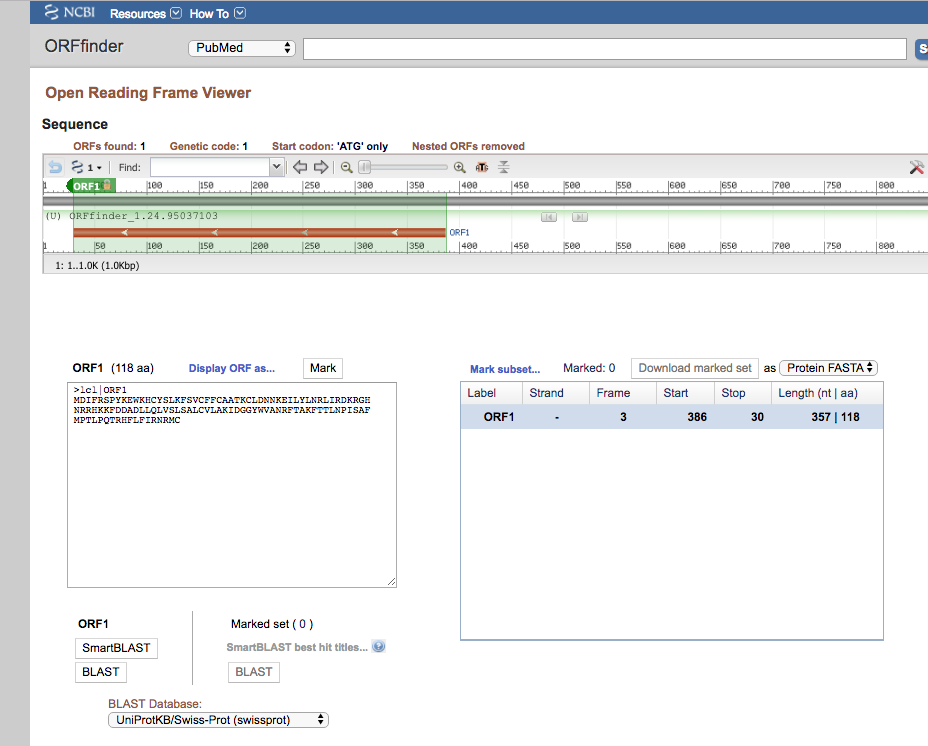
Figure 3. Screenshot, ORFfinder found ORFs.
3. If the ORF Finder Does Not Find ORFS
If no ORFs, then search the sequence for conserved domains. Conserved domains are functional units of a protein that act as building blocks in molecular evolution and recombine in various arrangements to make proteins with different functions. This website points to the Conserved Domain Database.
https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
Screenshot of the Conserved Domains site displayed in Fig 4.
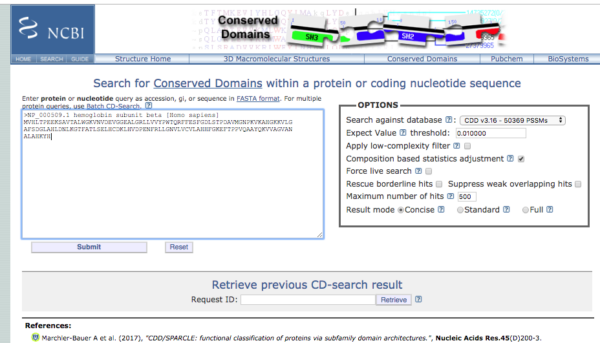
Figure 4. Screenshot homepage conserved domains search site at NCBI.
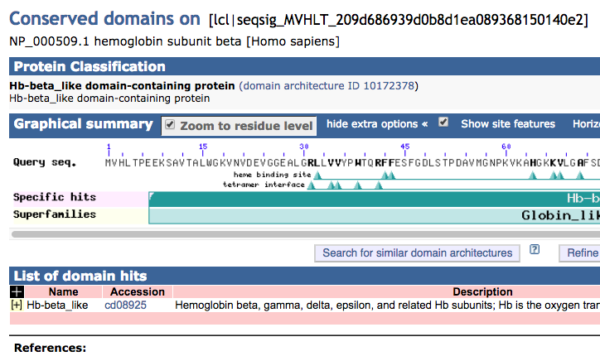
Figure 5. Screenshot of graphics results from Conserved Domains website for hemoglobin subunit beta.
Use of UGENE ORF finder menus.
1. Start UGENE.
2. Import sequence from file, or enter directly via File > New document from text...
Can import sequences one at a time or paste multiple sequences in FASTA format into the box. In this example, ten random sequences named Seq01 – Seq10 were imported.
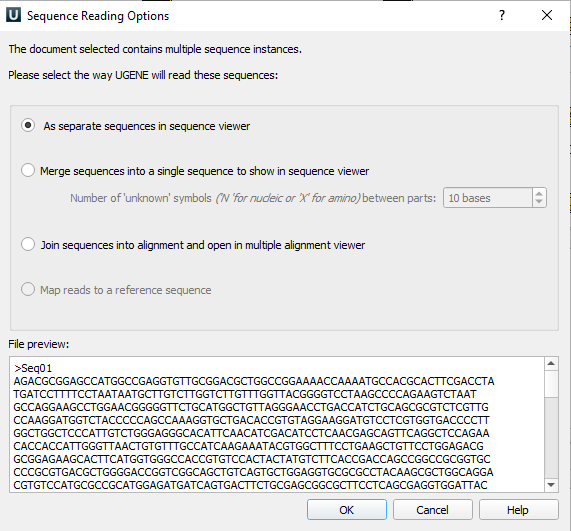
Figure 6. Screenshot of import sequence in UGENE.
One the sequence is accepted it will be visible in UGENE’s sequence window (Fig 7).
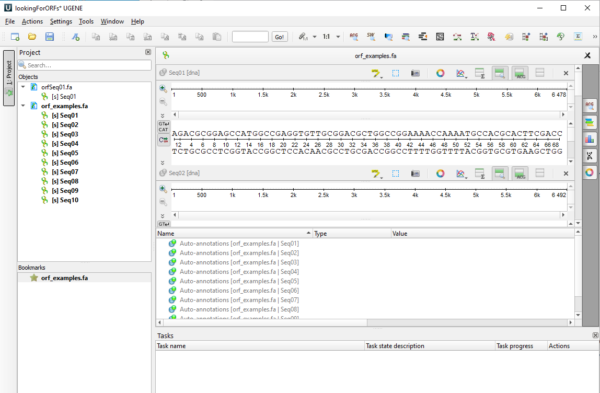
Figure 7. Screenshot of UGENE sequence window with imported DNA sequences.
3. Next, open Orf Maker. UGENE menu: Actions > Analyze > Find Orfs...Adjust settings or go with the defaults (Fig 8).
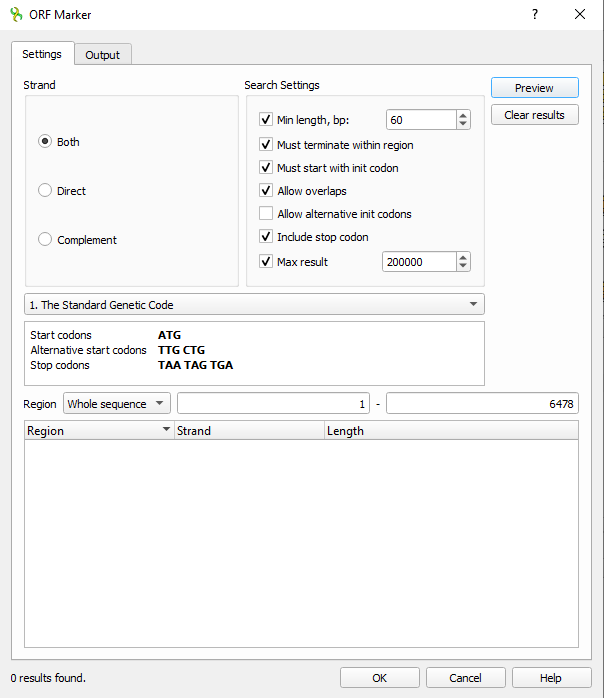
Figure 8. Screenshot UGENE ORF maker settings.
4. Before proceeding, click on the Output tab and enter information to save results to a text file (Fig 9).
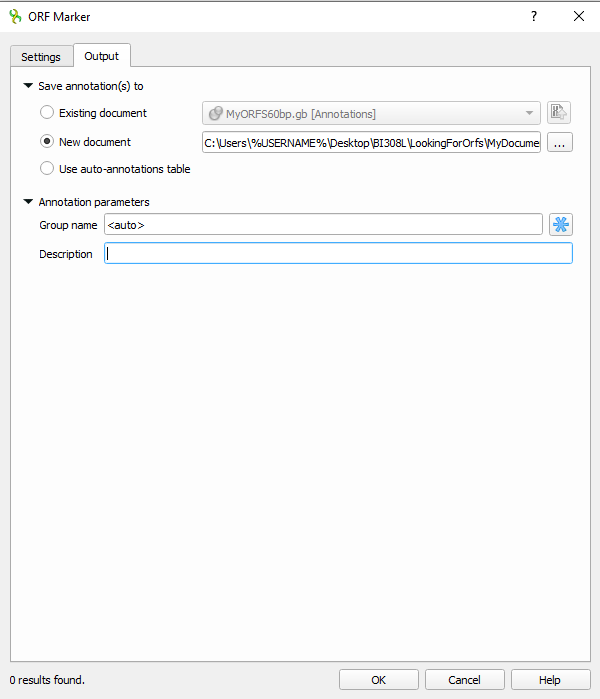
Figure 7. Screenshot UGENE ORF maker Output menu with file output information.
Once the settings are completed, click OK to proceed to search for ORFs.
5. Once ORF maker is complete, UGENE returns to the sequence viewer, now replete with annotations on your sequence(s) (Fig 8).
ORFs are noted in orange arrows. The direction of the arrows indicates the direction of translation among the six possible reading frames given a DNA sequence. Right-pointing arrows suggest translation along the forward strand, while left-pointing arrows suggest translation along the reverse strand. Of course, translation always proceeds 5-prime to 3-prime.
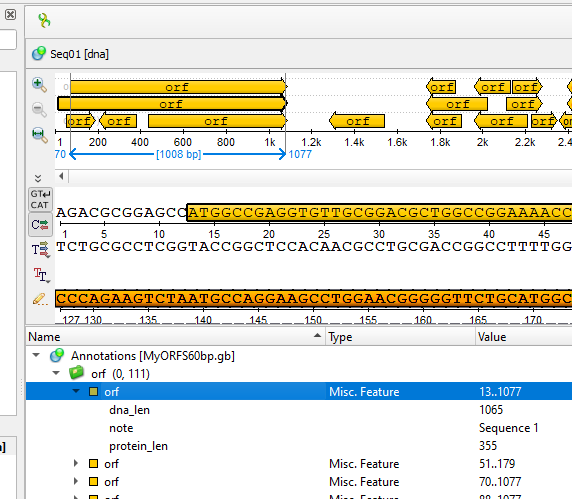
Figure 8. Screenshot UGENE ORF maker results displayed in sequence viewer.
6. Save the results.
Select one or more “orf” from the table displayed below the sequence viewer. For this example, I selected the first orf returned (Fig 8).
Go to Actions > Copy/Paste> Copy annotation amino acids
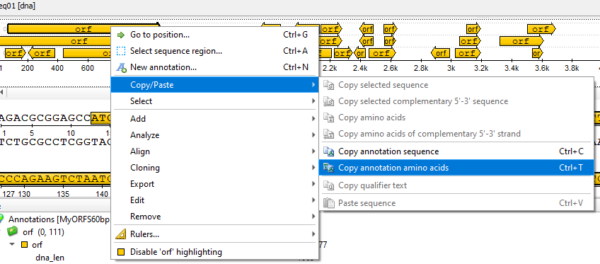
Figure 9. Screenshot UGENE menu selection to copy annotation results.
We summarize the results: eleven ORFs were identified, ranging in length from 42 to 183 nt. Putative proteins ranged in size from 13 to 60 amino acids.
Next steps could be to run BLAST on one or more of the ORFs to verify its function, identify mutations, and determine its relationship to other genes. We can select them one at a time by clicking on the subset table at lower right of the sequence viewer (eg, Fig 8).
What to do, what to turn in
- Generate your own random 1000 nt long DNA sequence at https://www.bioinformatics.org/sms2/random_dna.html
- You can use this sequence for Virtual Ribosome exercise, too, or generate a new one for each exercise
- Paste your sequence in FASTA format to ORF Finder, set ORF length to 30, then investigate for presence of ORFs.
- Copy and paste this same random sequence into the text submit box, no screenshots (so I can recreate your work).
- Capture a screenshot like the one in Figure 2. If no ORFs found, report as such.
- Select one ORF and do BLAST search. Report your results.
- Your responses to other questions in this handout should be placed in your notebook.