Using BLAST to predict PCR product length
to do:
- update images
- update links
Biological information is encoded in the nucleotide sequence of DNA. Bioinformatics is the field that identifies biological information in DNA using computer-based tools. Some bioinformatics algorithms aid the identification of genes, promoters, and other functional elements of DNA. We will use bioinformatic tools to investigate the location of several human short-tandem-repeat, STR, loci (assigned in class). In subsequent weeks we will obtain STR loci for samples of human DNA by PCR.
Upon completion of this exercise you will be able to identify
- The location in the genome where the STR locus is
- Make predictions about the outcome of your PCR experiment, that is, what size in numbers of base pairs is the PCR product (aka amplicon)?
Background — CODIS, the FBI’s DNA Databank
(this section needs updating — adapted from Arizona Biology Project)
The Federal Bureau of Investigation (FBI) of the US has been a leader in developing DNA typing technology for use in the identification of perpetrators of violent crime. In 1997, the FBI announced the selection of 13 STR loci to constitute the core of the United States national database, CODIS (Fig 1). All CODIS STRs are tetrameric repeat sequences. All forensic laboratories that use the CODIS system can contribute to a national database. DNA analysts like Bob Blackett can also attempt to match the DNA profile of crime scene evidence to DNA profiles already in the database.
There are many advantages to the CODIS STR system:
- The CODIS system has been widely adopted by forensic DNA analysts
- STR alleles can be rapidly determined using commercially available kits.
- STR alleles are discrete, and behave according to known principles of population genetics
- The data are digital, and therefore ideally suited for computer databases
- Laboratories worldwide are contributing to the analysis of STR allele frequency in different human populations
- STR profiles can be determined with very small amounts of DNA
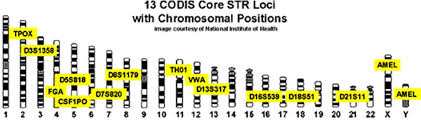
Figure 1. CODIS STR
A DNA Profile: The 13 CODIS STR loci
As part of his training and proficiency testing for DNA Profile analysis of STR (Short Tandem Repeat) Polymorphisms, Forensic Scientist and DNA Analyst Bob Blackett created a DNA profile on his own DNA. Here is Bob’s DNA Profile for the 13 core Genetic Loci of the United States national database, CODIS (Combined DNA Index System):
| Locus | D3S1358 | vWA | FGA | D8S1179 | D21S11 | D18S51 | D5S818 |
| Genotype | 15, 18 | 16, 16 | 19, 24 | 12, 13 | 29, 31 | 12, 13 | 11, 13 |
| Frequency | 8.2% | 4.4% | 1.7% | 9.9% | 2.3% | 4.3% | 13% |
| Locus | D13S317 | D7S820 | D16S539 | THO1 | TPOX | CSF1PO | AMEL |
| Genotype | 11, 11 | 10, 10 | 11, 11 | 9, 9.3 | 8, 8 | 11, 11 | X Y |
| Frequency | 1.2% | 6.3% | 9.5% | 9.6% | 3.52% | 7.2% | (Male) |
The complete primer set we have at Chaminade for STR is listed below.
| CUH Primer name | Locus designation | Primers (5′ – 3′) | Used for |
| CFamelS1 | Amelogenin | ACCTCATCCTGGGCACCCTGG | |
| CRamelS1 | Amelogenin | AGGCTTGAGGCCAACCATCAG | |
| CFcsfS1 | CSF1PO | AACCTGAGTCTGCCAAGGACTAGC | |
| CFcsfS2 | CSF1PO | CCGGAGGTAAAGGTGTCTTAAAGT | |
| CRcsfS1 | CSF1PO | TTCCACACACCACTGGCCATCTTC | |
| CRcsfS2 | CSF1PO | ATTTCCTGTGTCAGACCCTGTT | |
| CFd18sS1 | D18S51 | CAAACCCGACTACCAGCAAC | |
| CFd18sS2 | D18S51 | TTCTTGAGCCCAGAAGGTTA | Primer 3 F |
| CRd18sS1 | D18S51 | GAGCCATGTTCATGCCACTG | |
| CRd18sS2 | D18S51 | ATTCTACCAGCAACAACACAAATAAAC | Primer 3 R |
| CFd3sS1 | D3S1358 | ACTGCAGTCCAATCTGGGT | |
| CFd3sS2 | D3S1358 | ACTGCAGTCCAATCTGGGT | Primer 2 F |
| CRd3sS1 | D3S1358 | ATGAAATCAACAGAGGCTTG | |
| CRd3sS2 | D3S1358 | ATGAAATCAACAGAGGCTTGC | Primer 2 R |
| CFd7sS1 | D7S820 | ATGTTGGTCAGGCTGACTATG | |
| CFd7sS2 | D7S820 | TGTCATAGTTTAGAACGAACTAACG | |
| CRd7sS1 | D7S820 | GATTCCACATTTATCCTCATTGAC | |
| CRd7sS2 | D7S820 | CTGAGGTATCAAAAACTCAGAGG | |
| CFdysS1 | DYS391 | CTATTCATTCAATCATACACCCA | |
| CFdysS2 | DYS391 | TTCATCATACACCCATATCTGTC | |
| CRdysS1 | DYS391 | GATTCTTTGTGGTGGGTCTG | |
| CRdysS2 | DYS391 | GATAGAGGGATAGGTAGGCAGGC | |
| CFfgaS1 | FGA | ATTATCCAAAAGTCAAATGCCCCATAGG | |
| CFfgaS2 | FGA | GGCTGCAGGGCATAACATTA | |
| CRfgaS1 | FGA | ATCGAAAATATGGTTATTGAAGTAGCTG | |
| CRfgaS2 | FGA | ATTCTATGACTTTGCGCTTCAGGA | |
| CFthoS1 | TH01 | GTGGGCTGAAAAGCTCCCGATTAT | |
| CFthoS2 | TH01 | GCTTCCGAGTGCAGGTCACA | |
| CRthoS1 | TH01 | ATTCAAAGGGTATCTGGGCTCTGG | |
| CRthoS2 | TH01 | CAGCTGCCCTAGTCAGCAC | |
| CFtpS1 | TPOX | ACTGGCACAGAACAGGCACTTAGG | |
| CFtpS2 | TPOX | CACTAGCACCCAGAACCGTC | Primer 1 F |
| CRtpS1 | TPOX | GGAGGAACTGGGAACCACACAGGT | |
| CRtpS2 | TPOX | CCTTGTCAGCGTTTATTTGCC | Primer 1 R |
The CUH primer name reads as follows.
- C stands for Chaminade
- the F or R following C stands for forward (F) or reverse (R) primer sequence
- next three letters corresponds to first three letters of locus name
- S1 refers to primer set 1; S2 refers to primer set 2
To begin the Bioinformatics exercise, locate the primer set (forward, begins with “CF”, and reverse, begins with “CR”) from the Chaminade table. For example, if the CUH name is CFdysS1, then this is the forward primer for DYS391; the matching reverse primer then will be CRdysS1. Once you have the two primers for the STR locus, proceed to the next steps in the handout.
Conduct a BLAST search with your primer set
1. Go to https://www.ncbi.nlm.nih.gov/tools/primer-blast/ and copy exactly our primer sequences in the “Primer Parameters” area (look for “Use my own forward primer; Use my own reverse primer”). One at a time, copy and paste only the sequences, not the leading “5-” or trailing “-3’”. See red arrow, Fig 2.
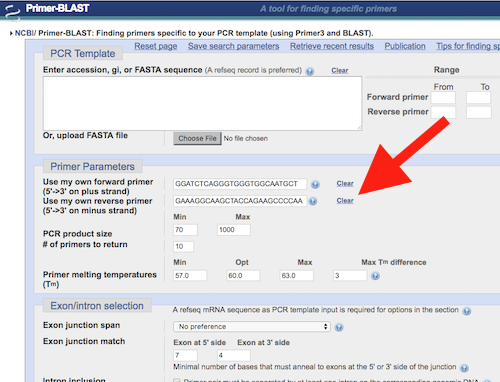
Figure 2.
2. While on the same page, scroll down to Database under “Primer Pair Specificity Checking Parameters” and change the default entry from “Refseq mRNA” to “Genome (reference sequence from selected organisms)”. Confirm that entry for Organism is “Homo Sapiens” or “9606”, the “taxid” for humans in the NCBI databases. See blue arrow next screenshot (Figure 3).
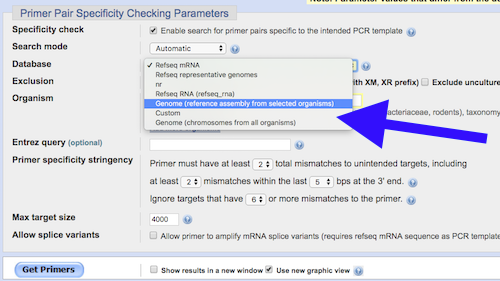
Figure 3.
3. Scroll down to find the “Get Primers” button (Fig 3). Click on the button – you should see a screen like this next one (Fig 4), which indicates the database search is proceeding.
Figure 4.
Question: What time zone is the database/server located? Hint: I conducted the search at 9:33AM HST.
4. After the search is complete (and successful!), you’ll get a screen like the one below (Fig 5).
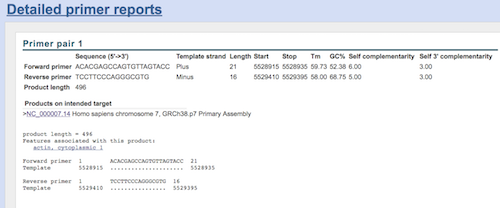
Figure 5.
Question. What is the predicted amplicon size?
This was an example for primer search for ACTB; You’ll get a similar page for your search. Click on the link under the “Products on intended target (e.g., NC_000007.14 in this example) to bring up the sequence page (Fig 6).
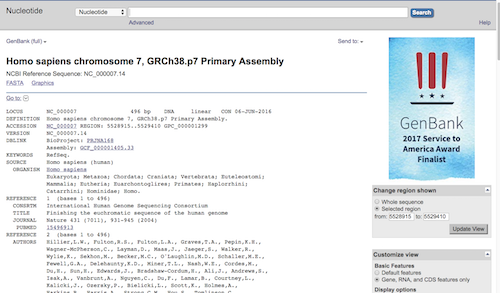
Figure 6.
Question. Where in the genome is your target STR sequence? Note not only the chromosome number, but the region in the genome, i.e., from base number: ____ to base number: _____.
/MD