An example consensus phylogenetic tree
A consensus tree is the representation of phylogenetic relationships that incorporates summarized information from a variety of sources, including different kinds of characters and tree building algorithms (Bryant et al 2017 and references therein). It can serve as a reference (Huson and Cetinkaya 2023 and references therein) to evaluate incompatibilities between gene trees, which reflect the evolutionary history of the gene vs the common relationships among a set of taxa used to infer a species tree.
This page describes approaches to obtain a consensus tree based on summaries from a variety of approaches to generate species trees. An alternative approach is described on another page in this manual for using the NICBI Taxonomy browser to generate a consensus tree.
See also How to read a phylogeny and Working with Newick file format.
Worked example
Our 13+1 taxa (species) list
Alligator
Cat
Cattle
Chicken
Chimpanzee
Dog
Human
Lizard
Macaque
Mouse
Opossum
Pig
Rabbit
Rat
Our consensus phylogeny was generated from timetree.org. The phylogenetic tree is shown in Fig 1A (with Newick string in Fig 1B).
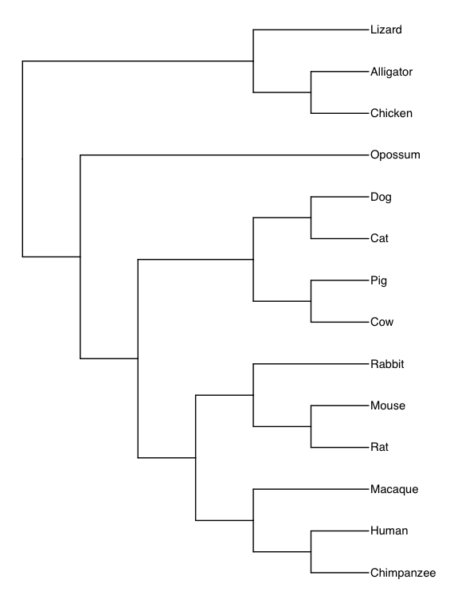
Figure 1A. Consensus phylogeny for our 14 species, compiled from data available at timetree.org. Branch lengths not displayed. Newick text in Figure 1B.
((Lizard:279.65697667,(Chicken:236.50266286,Alligator:236.50266286):43.15431381):32.24694470,(Opossum,(((Cat:54.32144118,Dog:54.32144118):23.43351523,(Cattle:61.96598852,Pig:61.96598852):15.78896789):18.70743276,((Rabbit:82.14079889,(Rat:20.88741740,Mouse:20.88741740):61.25338149):7.68238853,(Macaque:29.44154682,(Chimpanzee:6.65090500,Human:6.65090500):22.79064182):60.38164060):6.63920175):62.13519841):153.30633379);
Figure 1B. Newick code for tree in Figure 1A.
Update Spring 2022, changed Cow to Cattle in the Newick files on this page, but I did not updated the figures.
The branch lengths in the consensus tree (Fig 1A and 1B) are divergence times in millions of years (mya), and are not comparable to the gene tree branch lengths , which are genetic distances. You’ll need to remove the branch lengths before trying the reconciliation steps, which was done for you (Fig 2)
((Lizard,(Chicken,Alligator)),(Opossum,(((Cat,Dog),(Cattle,Pig)),((Rabbit,(Rat,Mouse)),(Macaque,(Chimpanzee,Human))))));
Figure 2. Newick code for consensus tree without divergence time branch lengths.
Considered together, do our genes trees agree with the consensus tree?
About consensus as an estimate of truth. When constructing a gene tree we are interested in the evolutionary history of that gene since the last common ancestor of our taxa. We may also be interested in reconstructing the evolutionary history of the taxa, but, because one gene is only one datum, a single point, we don’t expect a gene tree to agree completely with the true phylogeny for a collection of taxa. In principle, we do expect that adding additional information, i.e., additional data points, from many genes should converge on the true phylogeny.
There are two ways to develop a consensus tree from a set of protein (or DNA) sequences (Gadagkar et al 2005). One approach involves creating a new sequence by concatenating all protein (or DNA) sequences for each taxa in the dataset. The new combined sequences then needs to be aligned and the alignments used to build a tree. This we could do, and UGENE provides a way to concatenate sequences, but it’s quite a bit of work.
The second approach uses the gene trees we have already built; we simply look for agreement among a set of gene trees. And so, this is how to use R to do it. The results yielded the following tree (Fig 3).
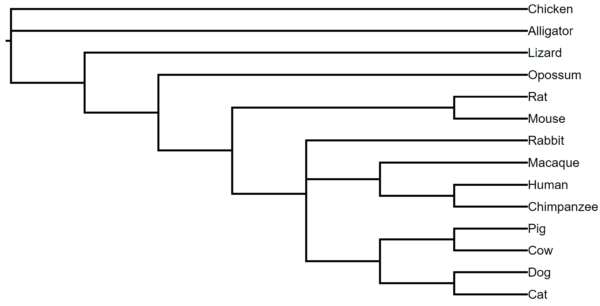
Figure 3. 2020 BI308L consensus tree based on 27 genes. Student gene trees were estimated by phyML approaches.
and the Newick code is
(((((((Cat,Dog),(Cattle,Pig)),((Chimpanzee,Human),Macaque),Rabbit),(Mouse,Rat)),Opossum),Lizard),Alligator,Chicken);
The next step would be to perform tree reconciliation, e.g., compare our timetree consensus tree versus the tree derived from consensus of all 27 genes. The result was a distance of 3: you should be able by now to identify the tree differences.
Method and R code for making the consensus tree from all 27 genes
First step, load all of the gene trees into R. The simplest way to do this is to create a NEXUS-formatted text file (Maddison et al. 1997). (NEXUS files can hold multiple trees). The NEXUS data for BI308L 2020 is listed after the References. Make sure the NEXUS file is in the working directory for R.
Next, copy and run the R code into R running on your computer (this code won’t work on rdrr.io)
#R code begins
library(ape)
library(phangorn)
library(phytools)
allTrees <- read.nexus("alltrees.nxs")
attach(allTrees)
#This next step gathers all of the gene names. The first tree in the file is the
#consensus tree from timetree.org is called agreeTree. We want the other gene trees.
studentTrees <- allTrees[2:27]
#values for p, which can be between 0 and 1. Higher values of p result in more
#polytomies, values of p too small and the tree won't plot.
tryAgree <- consensus(studentTrees,p = 0.3, check.labels = TRUE)
plot(tryAgree)
#Get the tree distance between the two consensus trees
RF.dist(agreeTree,tryAgree)
#save the new tree to a Newick file in the working directory
write.tree(tryAgree, file="BI308L_consensus.nxs")
#End R code
Finally, open the file BI308L_consensus.nxs in your text editor to retrieve the tree code.
References
Bryant, D., Francis, A., & Steel, M. (2017). Can We “Future-Proof” Consensus Trees?. Systematic Biology, 66(4), 611–619.
Gadagkar, S. R., Rosenberg, M. S., & Kumar, S. (2005). Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 304(1), 64-74.
Huson, D. H., & Cetinkaya, B. (2023). Visualizing incompatibilities in phylogenetic trees using consensus outlines. Frontiers in Bioinformatics, 3.
Maddison, D. R., Swofford, D. L. and Maddison, W. P. (1997) NEXUS: an extensible file format for systematic information. Systematic Biology, 46, 590–621.
NEXUS dataset
Copy from #NEXUS to end of page. Paste into new text file. Save as allTrees.nxs .
#NEXUS BEGIN TAXA: TAXALABELS Lizard Chicken Alligator Opossum Cat Dog Cattle Pig Rabbit Rat Mouse Macaque Chimpanzee Human; END; BEGIN TREES; TREE agreeTree = ((Lizard,(Chicken,Alligator)),(Opossum,(((Cat,Dog),(Cattle,Pig)),((Rabbit,(Rat,Mouse)),(Macaque,(Chimpanzee,Human)))))); TREE ZNF320 = (Alligator:0.0364011,Chicken:0.0394441,((((((Cat:0.00859765,Dog:0.00745599):0.00888461,(Cattle:0.00941794,Pig:0.00693555):0.0137144):0.00513651,((Chimpanzee:0.00096848,Human:0.00191374,Macaque:0.00536105):0.0179603,Rabbit:0.028247):0.00466631):0.0028666,(Mouse:0.00469363,Rat:0.00647799):0.0101139):0.0247477,Opossum:0.0179645):0.0514154,Lizard:0.0796556):0.0249907); TREE SERPINA9 = (Alligator:0.341987,Chicken:0.423977,(((((Cat:0.106657,Dog:0.113384):0.0970512,((Mouse:0.0900277,Rat:0.0504645):0.202323,Rabbit:0.222122):0.0706069):0.0351808,((Chimpanzee:0.0130481,Human:0.0157507):0.140082,Macaque:0.0572265):0.126478):0.251356,((Cattle:0.10814,Pig:0.0666009):0.28567,Opossum:0.379823):0.0672844):0.255245,Lizard:0.513237):0.108521); TREE MYO16 = (Human:0.0045964,Chimpanzee:0.00401286,(((((Mouse:0.0314074,Rat:0.029889):0.180268,Rabbit:0.173399):0.0306359,Lizard:29.5072):0.0299355,(((Alligator:0.0836772,Chicken:0.0900185):0.111966,Opossum:0.091326):0.10298,((Cat:0.0675349,Dog:0.099453):0.0484925,(Cattle:0.0990281,Pig:0.0966564):0.0596204):0.037329):0.0189681):0.0830799,Macaque:0.015677):0.0143454); TREE AHR = (Alligator:0.0664323,Chicken:0.1262,((((Cat:0.116644,(Cattle:0.10141,Pig:0.0776841):0.0242184):0.0205112,(((Chimpanzee:0.00519902,Human:0.0108034):0.02132,Macaque:0.0229029):0.0646343,((Mouse:0.0524342,Rat:0.0452807):0.239138,Rabbit:0.141519):0.0380129):0.0355584):0.121387,(Dog:20.3157,Opossum:0.43592):0.672323):0.175237,Lizard:0.18292):0.031559); TREE CYP2A6 = (Human:0.0274382,Macaque:0.0186449,(Alligator:3.14081,(Chicken:1.00871,Chimpanzee:0.00428747):0.760237):0.488739,((((((Lizard:0.475004,Opossum:0.0853667):0.0596331,(Mouse:0.0126904,Rat:0.0168983):0.0637912):0.013409,(Cat:0.0419809,Dog:0.02058):0.0185491):0.0122308,Pig:0.0414598):0.0097499,Cattle:0.0265408):0.0116579,Rabbit:0.0881735):0.0558017); TREE SLC17A3 = (Rabbit:0.158882,((((Rat:0.0686039,Mouse:0.0869711):0.172134,((Dog:0.119165,Cat:0.174484):0.058094,((Chimpanzee:0.00789996,Human:0.00541463):0.0451324,Macaque:0.0398237):0.128601,Cattle:0.200842):0.0787009,Alligator:5.38693):0.249736,Opossum:0.432174):0.0816892,Pig:0.454562):0.889829,(Chicken:0.0774528,Lizard:0.107805):0.0526092); TREE MICA = (Human:0.128848,Chimpanzee:0.070676,((((((Alligator:0.696092,Lizard:0.496513):0.134104,Chicken:0.552255):0.130692,((Cat:0.205751,Rabbit:0.368953):0.111118,(Mouse:0.208443,Rat:0.19111):0.119399):0.278392):0.152292,Dog:0.776837,Opossum:1.74439):0.607713,(Cattle:0.317675,Pig:0.413756):0.246383):0.256236,Macaque:0.12594):0.0641231); TREE MROH9 = (Rat:0.00153687,((((Alligator:0.0167929,Chicken:0.00147466):0.00730464,Lizard:0.169708):0.00797142,Opossum:0.0117042):0.0189034,(Cat:5e-06,(Chimpanzee:5e-06,(Cattle:5e-06,(Dog:5e-06,(Macaque:5e-06,(Pig:5e-06,(Rabbit:5e-06,Human:5e-06):0):0):0):0):0):0):0.000191698):0.00131875,Mouse:0.00149269); TREE OCA2 = (Alligator:0.164659,Chicken:0.285118,(Lizard:0.26755,(Opossum:0.160099,((Mouse:0.0354956,Rat:0.0346325)293.34:0.288334,(Rabbit:0.147037,((Macaque:0.040178,(Chimpanzee:0.00558464,Human:0.0126416)33.2223:0.0206436)292.046:0.135887,((Cat:0.0513166,Dog:0.0714231)162.259:0.0860443,(Cattle:0.0935381,Pig:0.12379)48.0609:0.0430212)62.9763:0.0502294)2.61918:0.00856966)5.41581:0.0368795)312.494:0.35017)103.198:0.161373)17.6715:0.0698452); TREE FBXO31 = (Alligator:0.447911,Lizard:0.681805,(Chicken:0.353718,(Opossum:0.300509,((Mouse:0.00467884,Rat:0.0604603)76.3098:0.193518,((Chimpanzee:2.9e-07,(Macaque:0.0344749,Human:1e-08)4.08165:0.00351271)58.0428:0.126997,((Cat:0.023275,(Dog:0.00555863,Rabbit:0.599637)115.712:0.118024)22.5406:0.0613603,(Pig:0.0903906,Cattle:0.227307)7.5533:0.0348575)59.301:0.164767)5.16178:0.0375047)21.8808:0.203972)110.415:0.599743)1.4268:0.0579636); TREE CNRIP1 = (Alligator:0.0882769,Chicken:0.0564552,(Lizard:0.10385,(Opossum:0.0249756,((Cat:0.0108017,(Chimpanzee:0.0046655,Human:0.00692616):0.0221197):0.0117957,Mouse:0.0190273):0.0324191):0.197098,(((Cattle:0.00705539,Pig:0.00354332):0.00612414,Dog:0.00285421):0.00330831,(Macaque:0.000859892,Rabbit:0.00233292):0.0114954,Rat:0.0124235):2.84683):0.0576188); TREE RXRG = (Chicken:0.177921,Alligator:0.0416502,((Lizard:0.180531,Rabbit:1.35043):0.563266,((((Mouse:0.035789,Rat:0.0210154):0.0139804,(((Cat:0.0344363,Dog:0.0544332):0.0328621,Pig:0.00884493):0.00499901,Cattle:0.0123239):0.00535919):0.00738245,(Human:0.00641145,Chimpanzee:0.00198076):0.00594774,Macaque:0.00351092):0.125568,Opossum:0.142236):0.0996573):0.0795474); TREE CERS6 = (Cat:0.034266,(Cattle:0.0410462,((((Alligator:0.099943,Chicken:0.0669836):0.0159057,Lizard:0.108026):0.110038,Opossum:0.0348357):0.0491857,(Pig:0.0165024,(((Human:5e-06,Chimpanzee:5e-06):0.00317074,Macaque:0.00222984):0.0108141,((Mouse:0.00412878,Rat:0.00671163):0.0274051,Rabbit:0.0228149):0.0021286):0.00481754):0.005448):0.00524473):0.0123647,Dog:0.0301371); TREE MAPT = (Alligator:0.0996946,(Lizard:0.158552,(Opossum:0.107973,((Mouse:0.00976438,Rat:0.0174102):0.0280738,((((Chimpanzee:5e-06,Human:5e-06):0.00949333,Macaque:0.0104683):0.0225772,Rabbit:0.052796):0.000235921,((Cattle:0.0389948,Pig:0.0121002):0.00245202,(Cat:0.0251882,Dog:0.0149863):0.00234487):0.0105962):0.0131822):0.0439099):0.0556089):0.020387,Chicken:0.0600185); TREE NECTIN2 = (Alligator:1.05203,Chicken:2.72e-05,(Lizard:0.00075785,(Opossum:1.23,((Rabbit:0.111517,(Mouse:0.0143255,Rat:0.025644)264.773:0.173713)4.97865:0.0550607,(Cat:0.00104766,(Dog:0.00733908,((Macaque:0.0246986,(Chimpanzee:0.00867818,Human:0.00347206)43.8974:0.023741)360.385:0.226628,(Cattle:0.130324,Pig:0.0871881)43.5081:0.0555787)62.4826:0.0889578)754.535:0.64944)4.78851:0.0479373)15.2078:0.528358)0.372836:2.14273)590.118:35.5201); TREE C10ORF32 = (Chimpanzee:0.0149519,Human:0.00458162,(Macaque:0.0149614,((((Alligator:0.0720781,(Chicken:0.0101931,Lizard:0.140871):0.0846644):0.0898932,Opossum:0.0651177):0.0963906,Cattle:0.0118556,Pig:0.00790228):0.0160869,(Mouse:0.0118304,Rat:0.0287091):0.0453917,Cat:0.00673568,Dog:0.0170335,Rabbit:0.0225397):0.0242659):0.0341791); TREE SRBD1 = (Chimpanzee:0.000848937,(Macaque:0.00336705,(Rabbit:0.0148956,(((Mouse:0.00680646,Rat:0.00557283):0.0456019,(((Alligator:0.0379122,Chicken:0.0353385):0.0114277,Lizard:0.0693876):0.0346713,Opossum:0.0357623):0.0166335):0.00337307,((Cat:0.00482974,Dog:0.014441):0.00332716,(Cattle:0.0135309,Pig:0.00953194):0.00190628):0.00627358):0.00294699):0.00788073):0.000340647,Human:0.00161526); TREE FTO = (Human:0.00144618,Chimpanzee:0.00468739,((((Mouse:0.023929,Rat:0.0200805):0.0915066,Rabbit:0.0377056):0.0247787,((((((Alligator:0.18683,Chicken:0.225813):0.0599907,Lizard:0.26188):0.227599,Opossum:0.269848):0.267585,Cattle:0.0636042):0.0189793,Pig:0.0556333):0.0100664,(Cat:0.0234945,Dog:0.0263813):0.0260214):0.0469853):0.0350907,Macaque:0.0102802):0.0115569); TREE TYR = (Alligator:0.0750033,Chicken:0.151716,(Lizard:0.140246,(Opossum:0.119399,(Rabbit:0.0629694,((Pig:0.0362316,(Cattle:0.0584014,(Cat:0.0534529,Dog:0.0273365):0.0267594):0.0110779):0.0261751,((Mouse:0.161034,Rat:0.0182243):0.0882978,(Macaque:0.0315679,(Human:0.00333246,Chimpanzee:0.0210227):0.0173107):0.0419984):0.0147291):0.0122723):0.0834216):0.108874):0.0540767); TREE CCRL2 = (((((Chimpanzee:8.61551,Macaque:-0.506127):3.56728,Lizard:0.567686):4.07771,Opossum:-1.03436):2.69909,Chicken:1.26749):1.00311,(Rat:2.73154,((((Cat:0.254656,Dog:0.192766):0.680685,(Cattle:0.160667,Pig:0.143462):0.898098):0.644046,Human:1.08238):0.543462,(Mouse:1.72615,Rabbit:0.337159):0.820927):0.867393):1.57946,Alligator:1.84704); TREE F2 = (Chimpanzee:0.00260148,(Macaque:0.0313039,((Mouse:0.0264288,Rat:0.041305):0.0993251,((((((Alligator:0.214771,Chicken:0.173341):0.0262897,(Lizard:0.16429,Dog:4.28844):0.0802896):0.113385,Opossum:0.251289):0.1037,Rabbit:0.166866):0.0126474,(Cattle:0.0990207,Pig:0.0678056):0.028887):0.00473998,Cat:0.0934627):0.00184045):0.0618306):0.0236414,Human:0.00236812); TREE GREB1 = (Chicken:0.0525214,Alligator:0.0472261,(Lizard:0.107292,(Opossum:0.0743766,(((Mouse:0.00224789,Rat:0.00303188):0.0825257,Rabbit:0.122054):0.0114083,(((Cat:0.0330939,Dog:0.0279618):0.0145144,(Cattle:0.0357639,Pig:0.050768):0.0191187):0.0163439,((Chimpanzee:0.00241153,Human:0.00604683):0.00659922,Macaque:0.00999443):0.0375456):0.0126293):0.115024):0.0695975):0.0353527); TREE CPSF2 = (Chicken:0.0111816,(Lizard:0.0268787,(Opossum:0.00403024,((Mouse:0.00635689,Rat:0.00266569):0.00853187,(Cattle:0.00152977,(((Chimpanzee:0.00012496,Human:-0.00011496):0.00127473,Macaque:5.01358e-05):0.00113423,(Cat:0.00263337,((Dog:0.000822394,Rabbit:0.0030348):0.000325972,Pig:0.00224643):0.000587055):0.000222547):0.000319867):0.00161609):0.00349272):0.0132456):0.00579539,Alligator:0.00684141); TREE SLC8A1 = (Alligator:0.0816629,Chicken:0.124912,(Lizard:0.232993,(Opossum:0.140374,((Mouse:0.0213346,Rat:0.0279451)94.9222:0.0856717,(Pig:0.04719,((Cat:0.0192815,Dog:0.0281746)20.2633:0.019572,(Rabbit:0.130913,(Cattle:0.0646849,(Macaque:0.00536787,(Human:0.00786401,Chimpanzee:1e-06)14.3697:0.00844452)52.2483:0.0225524)0.375137:0.0030836)0:3.5e-07)0.374123:0.0135412)4.24436:0.00705058)25.8378:0.0479033)78.8698:0.11956)17.2451:0.0565737); TREE FGF5 = (Mouse:5.72e-06,Rat:0.117453,(((Chimpanzee:5.2e-07,(Human:3.7e-07,Macaque:0.0228347)1.15224:0.00377208)33.7113:1.89551,(Rabbit:2.88689,((Cattle:0.0348622,(Cat:0.0155137,Dog:0.0649193)42.7902:0.0430944)0.591832:0.0225066,((Lizard:13.0394,Pig:0.0251149)0.003862:0.0003754,Opossum:0.255182)14.1152:0.143164)5.87289:1.34848)1.666:0.489264)1.62394:0.71042,(Alligator:6.95164,Chicken:7.3654)1.12981:2.06477)5.5458:1.97404); TREE DAG1 = (Alligator:0.0587036,(Lizard:0.133506,(Opossum:0.0519929,((Mouse:0.00695824,Rat:0.0117307):0.0255042,(Rabbit:0.0337507,(((Cat:0.0160751,Dog:0.0227973):0.00605321,((Chimpanzee:0.00152062,Human:0.00394348):0.00598254,Macaque:0.0128429):0.0125557):0.000544026,(Cattle:0.0390797,Pig:0.0217691):0.00499464):0.00509669):0.00244433):0.0692833):0.0528855):0.0163573,Chicken:0.0691575); END;